Page 50 - ICAR-Accreditation-VIT-VAIAL SSR
P. 50
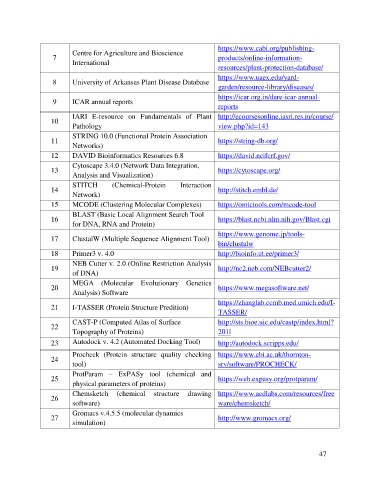
https://www.cabi.org/publishing-
Centre for Agriculture and Bioscience
7 products/online-information-
International
resources/plant-protection-database/
https://www.uaex.edu/yard-
8 University of Arkansas Plant Disease Database
garden/resource-library/diseases/
https://icar.org.in/dare-icar-annual-
9 ICAR annual reports
reports
IARI E-resource on Fundamentals of Plant http://ecoursesonline.iasri.res.in/course/
10
Pathology view.php?id=143
STRING 10.0 (Functional Protein Association
11 https://string-db.org/
Networks)
12 DAVID Bioinformatics Resources 6.8 https://david.ncifcrf.gov/
Cytoscape 3.4.0 (Network Data Integration,
13 https://cytoscape.org/
Analysis and Visualization)
STITCH (Chemical-Protein Interaction
14 http://stitch.embl.de/
Network)
15 MCODE (Clustering Molecular Complexes) https://omictools.com/mcode-tool
BLAST (Basic Local Alignment Search Tool
16 https://blast.ncbi.nlm.nih.gov/Blast.cgi
for DNA, RNA and Protein)
https://www.genome.jp/tools-
17 ClustalW (Multiple Sequence Alignment Tool)
bin/clustalw
18 Primer3 v. 4.0 http://bioinfo.ut.ee/primer3/
NEB Cutter v. 2.0 (Online Restriction Analysis
19 http://nc2.neb.com/NEBcutter2/
of DNA)
MEGA (Molecular Evolutionary Genetics
20 https://www.megasoftware.net/
Analysis) Software
https://zhanglab.ccmb.med.umich.edu/I-
21 I-TASSER (Protein Structure Predition)
TASSER/
CAST-P (Computed Atlas of Surface http://sts.bioe.uic.edu/castp/index.html?
22
Topography of Proteins) 201l
23 Autodock v. 4.2 (Automated Docking Tool) http://autodock.scripps.edu/
Procheck (Protein structure quality checking https://www.ebi.ac.uk/thornton-
24
tool) srv/software/PROCHECK/
ProtParam – ExPASy tool (chemical and
25 https://web.expasy.org/protparam/
physical parameters of proteins)
Chemsketch (chemical structure drawing https://www.acdlabs.com/resources/free
26
software) ware/chemsketch/
Gromacs v.4.5.5 (molecular dynamics
27 http://www.gromacs.org/
simulation)
47